
At the recent mRNA Conference in Szeged, geneticist Tamás Kiss captivated the audience with his presentation on the long-overlooked role of small regulatory RNAs. His latest research reveals that the Epstein-Barr virus, known for causing various types of tumors, uses its own snoRNAs to orchestrate site-specific chemical modifications of the ribosomal RNA in human host cells, taking control of protein synthesis within infected cells. How does a virus obtain small RNAs typically characteristic of higher organisms? What is the role played within cells by these small RNAs, originating from seemingly nonfunctional gene segments? How did Tamás Kiss realize that what he initially believed to be RNA debris in an experiment conducted ten years before the discovery of the microRNA was actually the type of small RNA that was central to this year’s Nobel Prize-winning research? What guidance did he offer his former colleague at the Szeged Biological Research Center, Katalin Karikó, who was then on the verge of her future Nobel Prize-winning discovery? Our interview with Tamás Kiss revisits fundamental genetic concepts while uncovering the remarkably fascinating story of RNA, the molecule that scripts the narrative of life.
All living organisms produce proteins through ribonucleic acid (RNA) molecules, with messenger RNAs (mRNAs) transporting genetic information from DNA to the ribosomes, where proteins are synthesized. Furthermore, ribosomal RNAs (rRNAs) serve as essential components of ribosomes, while transfer RNAs (tRNAs) facilitate the transport of necessary amino acids, both being directly involved in protein synthesis. Scientists have only recently discovered that, alongside these three types of RNA, a variety of small regulatory RNAs are also produced within cells, with many of them guiding site-specific modifications of other RNAs. Dr. Tamás Kiss, a geneticist and alumnus of the University of Szeged, former researcher at the Biological Research Center in Szeged, and currently a professor at the University of Toulouse, has gained international recognition for his groundbreaking research uncovering this important biological role.
A central dogma in biology
When a cell requires a specific protein, it transcribes the relevant segment of genetic code from DNA in the nucleus into various RNA molecules. These transcribed RNAs then undergo a series of maturation steps, during which small regulatory RNAs – once shrouded in mystery – contribute to the modification of ribonucleotides (the fundamental units of RNA). One of the most prevalent modifications is the conversion of the RNA nucleotide uridine into pseudouridine. This natural modification mirrors the artificial alteration made by Katalin Karikó to the messenger RNA, designed to circumvent an excessive immune response coming from the body. Once matured, the RNA exits the nucleus and enters the cytoplasm, where protein synthesis occurs. This sequence of events, which represents the flow of genetic information from DNA to RNA and then to proteins, is known as the central dogma of molecular biology.
Q: Professor, I would like to gain some insight into the role that the small regulatory RNAs you study play in the fundamental biological processes occurring within the cell. Can you begin by describing the environment within the cell nucleus where RNA synthesis and maturation take place?
A: Picture a bacterium: its cell essentially functions like a bag, with all the components inside mixed together. In contrast, eukaryotic cells, with their defined nuclei, exhibit a well-organized structure where everything is distinctly compartmentalized. For instance, nested within the nucleus is the nucleolus, an organelle that consists of a membraneless assembly of molecules. This is where ribosomal RNA (rRNA) is synthesized, matured, and assembled with nearly 80 ribosomal proteins to form functional ribosomes. The rRNA is initially transcribed as a long precursor rRNA containing the sequentially embedded 18S, 5.8S, and 28S rRNAs which are subsequently excised through complex enzymatic maturation processes. Meanwhile, over two hundred meticulously selected nucleotides undergo covalent modifications before the rRNA is incorporated into the small and large subunits of the ribosome.
Q: What evolutionary reason accounts for why the cell does not use RNAs in their initially transcribed form but, instead, requires them to undergo maturation?
A: Primary RNA products synthesized in the nucleus contain external and internal “non-coding” segments of varying lengths, which must be removed during RNA maturation to produce functional RNAs. For example, the coding sequences (exons) of precursor mRNAs generated during the transcription of protein-coding genes are interspersed with longer or shorter non-coding sequences (introns). During mRNA maturation, the intron regions are excised, and the ends of the exons are joined together in a process known as splicing, or cutting and joining. Occasionally, certain exons are omitted, while others are included in a new version of the mRNA – a process known as alternative splicing – which allows multiple proteins to be generated from a single gene, significantly enhancing the structural and functional diversity of the cell’s protein repertoire.
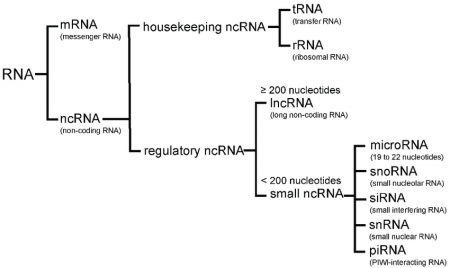
The categories of small RNAs. Source: Research Gate
Q: With your research group in Toulouse being the first to describe the function of snoRNAs, can you explain the role they play in this process?
A: Maturation is a process characteristic of RNAs in general; it is not limited to rRNA, as almost all RNAs undergo maturation and site-specific covalent nucleotide modifications. Approximately 5-6 percent of all RNA ribonucleotides found in human cells are modified, with the two most common modifications being the methylation of the 2’-hydroxyl group of ribose molecules and the conversion of certain uridine bases into pseudouridine. In other words, RNAs are typically not transcribed from DNA in the form in which they later exist and function in the cell.
By the end of the 20th century, much had been learned about the enzymatic cleavage and maturation of precursor RNAs; however, the site-specific synthesis of modified nucleotides remained one of the great mysteries in biology. My research group uncovered that the ribose methylation and pseudouridylation of rRNAs, small nuclear RNAs (snRNAs), and certain transfer RNAs (tRNAs) are regulated by small nucleolar RNAs (snoRNAs). These snoRNAs, responsible for rRNA modifications, are predominantly found in the nucleolus, hence the name small nucleolar RNA.
Once the small and large ribosomal subunits are fully formed during maturation, they migrate into the cytoplasm through the pores of the nuclear membrane, where they come together to form functional ribosomes. Following the genetic instructions carried by mRNAs, these ribosomes synthesize proteins. Thus, modified nucleotides are essential for the correct and efficient functioning of modified RNAs, playing a vital role in protein synthesis in the cytoplasm and in every step of gene expression taking place in the nucleus.
Q: How is it possible that, amid this diverse array of RNAs, proteins, and enzymes, it is a relatively small number of components that fulfill such a broad range of functions?
A: SnoRNAs are, in fact, guiding RNAs that identify the nucleotides to be modified. The snoRNA base-pairs with the complementary target sequence of the specific RNA, and then the RNA-modifying enzyme associated with the snoRNA executes the modification of the selected nucleotides, which often involves pseudouridylation or ribose methylation. Through this mechanism, a single enzyme, aided by hundreds of snoRNAs, can carry out a multitude of nucleotide modifications within the cell’s RNAs, eliminating the need to have hundreds of site-specific RNA-modifying enzymes evolve over time, a task that would be incredibly cumbersome. Instead, the guiding RNA’s target-recognition (antisense) sequences can readily accumulate sufficient point mutations to form a new recognition sequence, thereby resulting in the creation of a new snoRNA. In bacteria, each nucleotide modification is carried out by a specialized protein enzyme; and while bacterial RNAs typically contain far fewer modified nucleotides than their eukaryotic counterparts, approximately 7-8 percent of the bacterial genome still encodes RNA modification enzymes. Without snoRNAs, the number of genes responsible for RNA modifications in humans would be exponentially higher than it is today. It is also important to note that certain variants of snoRNAs that regulate RNA modification are involved in other processes as well. For instance, they facilitate the precise and efficient excision of rRNAs from precursor rRNAs and guide the synthesis of telomeric DNA at chromosome ends – thereby regulating the lifespan of healthy cells or stimulating the proliferation of cancer cells.

Dr. Tamás Kiss gives an interview at the mRNA conference, Szeged. Photo: Ádám Kovács-Jerney
Q: Do you subscribe to the so-called RNA World Hypothesis, which suggests that in a very early phase of the evolution of life – before the emergence of DNA and the translation of proteins – RNA molecules served as the sole means of information storage?
A: Yes, I do, as this is a thoroughly logical and widely accepted theory, particularly since RNA is the only biomolecule capable of both catalyzing enzymatic reactions and storing genetic information.
Q: Are there types of RNA with roles that are still unknown or types of RNA that are entirely undiscovered?
A: There are numerous non-coding RNAs (ncRNAs) in human cells. Among these, probably only a few small RNAs remain undiscovered, but many long RNAs might still be unknown. Long non-coding RNAs (lncRNAs) resemble traditional messenger RNAs, with their 3'-end poly(A) sequences and 5'-end caps. However, their protein-coding capacity is limited, as they lack extensive open reading frames for protein synthesis. Nevertheless, this does not rule out the possibility that short oligopeptides could be transcribed from lncRNAs, which are primarily linked to specific proteins in the cell and perform a variety of regulatory functions. For instance, the Xist lncRNA silences the transcription of one of the X chromosomes in females, effectively preventing undesirable chromosomal dosage effects. Additionally, several other lncRNAs are involved in regulating the transcription of specific chromosomal regions. Furthermore, some lncRNAs support protein activity, intracellular localization, or the maturation of other RNAs, while many of them regulate the formation and maintenance of certain cell organelle structures.
Q: Do you see any unresolved issues in the field of RNA research?
A: Although we discovered the role of snoRNAs in the maturation of ribosomal rRNA back in the 1990s, several intriguing details remain to be explored, since RNA is an extraordinary molecule – remarkably dynamic and capable of swiftly adapting its three-dimensional structure. My group recently discovered that snoRNAs, which guide RNA modifications, can form dynamic alternative structures with target sequences, thereby facilitating the specific modification of multiple target nucleotides. In addition, RNA can catalyze biochemical reactions, much like a classical protein enzyme. For example, the ribosome responsible for protein synthesis is, in fact, an RNA enzyme, with ribosomal proteins primarily supporting rRNA activity. Protein synthesis, therefore, is a process fundamentally catalyzed by RNA, as is the splicing of rRNAs, which is facilitated by small nuclear RNAs.
Q: Katalin Karikó often mentions in her presentations that your RNA research inspired her groundbreaking work on modified mRNA. In this interview, you’ve already discussed the transformation of the uridine nucleotide into pseudouridine – the very modification Professor Karikó ingeniously applied to messenger RNA to bypass excessive immune responses. Could this natural process have been the ‘inspiration’ for her later Nobel Prize-winning discovery?
A: If I recall correctly, the significance of modified nucleotides in preventing RNA-induced inflammation became apparent when Kati [Professor Karikó] and her team discovered that transfer RNA (tRNA), unlike other RNAs, did not trigger an immune response when injected into tissues. Since tRNA is the most heavily modified RNA in the cell and pseudouridine is the most common modified nucleotide in it, it was logical to assume that the modifications or the pseudouridine might play a role in avoiding an aggressive immune reaction.
Kati reached out to me to inquire whether I had any enzyme capable of converting all uridines in an RNA molecule to pseudouridine, but unfortunately, I had to tell her that no such enzyme had been discovered yet and that it probably does not exist. Indeed, our snoRNA/protein complexes directing pseudouridylation can only convert one specific uridine within the RNA.
Eventually, I proposed that we transcribe test mRNAs using T3, T7, or SP6 bacteriophage RNA polymerases in the presence of pseudouridine triphosphate. While this was a standard procedure for producing RNAs in test tubes, I was uncertain whether phage RNA polymerases would accept pseudouridine triphosphate as a substitute for uridine triphosphate.

Photo: Ádám Kovács-Jerney
Q: It was intriguing to learn from your presentation that the Epstein-Barr virus’s own snoRNA can direct the ribose methylation of the host’s ribosomal RNA, thereby modifying protein synthesis in the infected cell. How does this process work?
A: Rather than taking over translation, the virus modifies the translational system of the human host cell, with its viral snoRNA altering the function of the human ribosome in the infected cells by modifying ribosomal rRNA. While we have yet to grasp why this benefits the virus, we are pursuing promising avenues of research in collaboration with a viral expert, given that our own background is in molecular biology. To date, we have been studying a cell model – specifically, a cancer cell that expresses the complete genome of the Epstein-Barr virus. However, due to safety concerns, we have been unable to work directly with the virus at our institute.
Q: Does the virus’s genome also contain its own snoRNAs? What is the evolutionary history of this?
A: The virus contains only one snoRNA, which is clearly of human origin. Human snoRNA genes are located within the intron regions of protein-coding genes, where snoRNAs are transcribed as part of the precursor mRNAs of the host genes and subsequently excised from them. The snoRNA sequence holds all the essential information required for precise excision, efficient accumulation, and RNA modification. We hypothesize that a human snoRNA gene was integrated into the viral genome, leading to the emergence of a target sequence recognition element through spontaneous point mutations, which subsequently modified the host ribosome in a manner favorable for the virus. Viruses typically carry only the most essential genetic information due to their limited genome size, as the viral DNA must fit within the viral capsid, preventing the accommodation of a larger genome. The Epstein-Barr virus can accommodate approximately 172 kilobases of DNA, and whatever is in it must be essential. In contrast, the human genome is considerably larger and is filled with various remnants from evolution. It contains many sequences that appear to serve no purpose – or have a purpose that remains unknown to us.
Q: In your presentation, you also shed light on other areas of cancer research where a deeper understanding of snoRNAs could be pivotal. Can snoRNA be a biomarker for predicting specific types of cancer?
A: Over the past decade, dozens of snoRNAs have been linked to the development and progression of a variety of cancer types. Researchers are sequencing a multitude of RNA samples from cancer cells, carefully analyzing this vast amount of data to determine whether the accumulation of snoRNAs varies across different cancer types – specifically, whether their numbers increase or decrease significantly depending on the type of cancer involved. An elevated snoRNA level may suggest that the snoRNA involved promotes cancer progression; and distinct patterns of snoRNA accumulation have been described for various cancer types. However, the molecular basis of the role snoRNAs play in cancer remains completely unknown – although it is presumably related to the RNA modifications they regulate.
Q: This year’s Nobel Prizes were awarded in recognition of the discovery of microRNAs (miRNAs). It is said that your research played a crucial role in advancing the identification of miRNAs.
A: When the first paper describing microRNAs was published in Science, it so happened that I was the reviewer of the article. I even considered shifting my focus to miRNA research; however, with numerous ongoing projects, limited funding, and only a few collaborators, I ultimately chose not to move forward with it. Interestingly, our research group had actually encountered miRNAs previously – without recognizing what they were. What happened was that while cloning human snoRNAs, we used an enzyme called RNA ligase to attach a short artificial RNA molecule to both ends of the RNAs derived from the nucleolus, and we frequently observed that an unidentified sequence of approximately 20–22 nucleotides was nestled between the snoRNA and the attached artificial RNA. It almost seemed as if it had come from the Moon, since it was absent from any snoRNA gene the sequence of which was known to us. Unfortunately, at that time, the human genome sequence was only partially known, leading us to suspect that these small RNA molecules might be intermediates in the natural process of RNA degradation. Later, when miRNAs were described in 1993, we revisited our previous findings and confirmed that we had indeed observed the most common human microRNAs a decade before their actual discovery.

Geneticist Dr. Tamás Kiss. Photo: Ádám Kovács-Jerney.
Q: Did you find it frustrating?
A: It wasn’t particularly frustrating because, to be honest, we had no idea what we were seeing. It didn’t even cross our minds that we were uncovering biologically relevant RNAs. After all, during my time at university, we were taught that there were no RNAs shorter than transfer RNAs in the cell – which, as it turns out, is not true.
Q: How did your work at the Biological Research Centre in Szeged differ from the experiences you later had in Basel and Toulouse?
A: First of all, at the Biological Research Center in Szeged, we had to solve everything ourselves, which, in turn, meant that we were familiar with every detail of our research areas and the experimental methods we employed. In contrast, researchers in the West could simply consult a catalog and order the necessary reagents and enzymes. We, on the other hand, were allowed to order chemicals from abroad for hard currency only once a year. In fact, many times, I had to purify specific enzymes myself. For example, if we needed tobacco acidic pyrophosphatase enzyme to cleave the 5'-end cap of RNAs, we were supposed to order it from abroad for hard currency, but instead of doing so, we’d go down to the greenhouse, ask for a tray of tobacco, and just start purifying it. If it didn’t work out, we had to start over – but this process allowed us to truly understand what we were doing. In Western Europe, however, researchers already used specific kits for their work; they would just follow the protocol, adding two microliters from this tube and one microliter from the other one. This meant that PhD students were unaware of what they were doing or what was actually happening in the reaction tubes. By comparison, I prepared molecular-biology grade reagents myself, employing labor-intensive purification processes, using chemicals and enzymes of questionable purity provided by a local chemical supplies company. I worked incredibly hard, but I learned a lot from it. It is also true that I certainly lost a tremendous amount of time, which proved to be a loss in terms of results as well – considering that researchers who simply took the perfect reagents off the shelf or out of the fridge advanced much faster than those who had to do the purifying themselves. Still, the challenging research conditions in Szeged empowered me to design new protocols in molecular biology rather than just relying on existing ones.
Biography
Tamás Kiss serves as the Research Director of the French medical research organization INSERM, is an elected member of the European Molecular Biology Organization (EMBO) and holds a doctorate from the Hungarian Academy of Sciences. He graduated from the Faculty of Science at the University of Szeged and completed both his undergraduate and postgraduate training at the Szeged Biological Research Center of the Hungarian Academy of Sciences, where, in collaboration with Professor Ferenc Solymosy, he discovered and described small nuclear RNAs (snRNAs) in plants. His postdoctoral research in the Friedrich Miescher Institute in Basel, conducted in collaboration with Professor Witold Filipowicz, focused on understanding the RNA polymerase selection of plant snRNA genes. He was among the first to discover human small nucleolar RNAs (snoRNAs) encoded by introns. Later, as a group leader at the Laboratoire de Biologie Moléculaire Eucaryote at CNRS in Toulouse, he was the first to discover and describe several human small nuclear RNAs. He has elucidated the functions of several newly discovered and previously known small nuclear RNAs, identifying their roles in human gene expression. He was the one to demonstrate that intronic snoRNAs guide the 2'-O-methylation and pseudouridylation of rRNAs. He has also described a new class of RNAs that accumulate in Cajal bodies and govern the direct modification of spliceosomal snRNAs.
Original Hungarian text by Sándor Panek
Photos by Ádám Kovács-Jerney

